Mostofa Rafid Uddin
I am a doctoral student in the CPCB
Ph.D. Program in Carnegie Mellon University School of Computer Science, working with Dr. Min Xu. I am also a Center for Machine Learning and Health (CMLH) Fellow
in Digital Health Innovation.
My research interests include un/self-supervised learning, Representation Learning, Computational Biology,
3D Computer Vision, Explainable AI, Vision Foundation Models.
My PhD thesis involves developing unsupervised algorithms for modelling subcellular object morphology across scales and modalities.
Before joining my PhD, I graduated with B.Sc.Engg. in Computer Science and
Engineering from Bangladesh University of Engineering and
Technology(BUET) and later worked as a lecturer. During that time, I worked with Dr. Md. Shamsuzzoha
Bayzid on leveraging machine translation for protein structure prediction.
- First authored work "DiLO: Disentangled Latent Optimization for Learning Shape and Deformation in Grouped Deforming 3D Objects" got accepted at AAAI-2026 main track! (acceptance rate: 17.6% out of 23,680)
- First authored work "Localization of Macromolecules in Crowded Cellular Cryo-electron Tomograms from Extremely Sparse Labels" got accepted at Briefings in Bioinformatics!
- First authored work "Unsupervised Identification of Protein Compositions and Conformations via Implicit Content-Transformation Disentanglement" got accepted at ICCV 2025!
- Our XAI work "DiffCAM: Data-Driven Saliency Maps by Capturing Feature Differences" accepted at CVPR 2025 as Highlight!
- Received Outstanding Research Accomplishment Award from my PhD Program!!
- Received the prestigious CMLH fellowship in digital health for 2023!
- Our consortium project on BrCa tumor landscape got accepted at Nature Cancer!!
Publications
Please refer to my Google Scholar page for an up-to-date list with citations.
-
Mostofa Rafid Uddin, Jana Armouti and Min Xu. Unsupervised Identification of Protein Compositions and Conformations via Implicit Content-Transformation Disentanglement In Proceedings of International Conference on Computer Vision (ICCV), 2025.
-
Xingjian Li, Mostofa Rafid Uddin, et al. DiffCAM: Data-Driven Saliency Maps by Capturing Feature Differences In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2025. (Highlight)
-
Mostofa Rafid Uddin, Gregory Howe, Xiangrui Zeng, and Min Xu. Harmony: A Generic Unsupervised Approach for Disentangling Semantic Content From Parameterized Transformations In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2022, pp. 20646-20655. [News link]
-
Mostofa Rafid Uddin, et al. Localization of Macromolecules in Crowded Cellular Cryo-electron Tomograms from Extremely Sparse Labels. Briefings in Bioinformatics (Accepted), 2025.
-
Mostofa Rafid Uddin, et al. DiLO: Disentangled Latent Optimization for Learning Shape and Deformation in Grouped Deforming 3D Objects AAAI Conference on Artificial Intelligence (Accepted), 2026.
-
Mostofa Rafid Uddin, Thanh-Huy Nguyen, HM Shadman Tabib, Kashish Gandhi, Min Xu. Unsupervised Multi-scale Segmentation of Cellular cryo-electron Tomograms with Stable Diffusion Foundation Model Biorxiv, 2025.
-
Mostofa Rafid Uddin, et al. Deep Disentangled Representation Learning Reveals Neuron Subtype-Specific Nuclear Morphologies Across Aging in Mice and Humans. Briefings in Bioinformatics (Accepted), 2025.
-
Mostofa Rafid Uddin, Sazan Mahbub, Md Saifur Rahman, Md Shamsuzzoha Bayzid. SAINT: Self-Attention Augmented Inception-Inside-Inception Network Improves Protein Secondary Structure Prediction . Bioinformatics , Volume 36, Issue 17, 2020, Pages 4599-4608.
-
Sayali Onkar, Mostofa Rafid Uddin, et al. Immune landscape in invasive ductal and lobular breast cancer reveals a divergent macrophage-driven microenvironment . Nature Cancer. 2023. 4(4), 516-534.
-
Mostofa Rafid Uddin, et al. TomoPicker: Annotation-Efficient Particle Picking in Cryo-electron Tomograms. NeurIPS Workshop on Machine Learning for Structural Biology, 2024.
-
Mostofa Rafid Uddin et al. Feature detection in cryo-electron tomography image analysis .Cryo-electron Tomography. Academic Press , 2025. 173-215.
Research Details
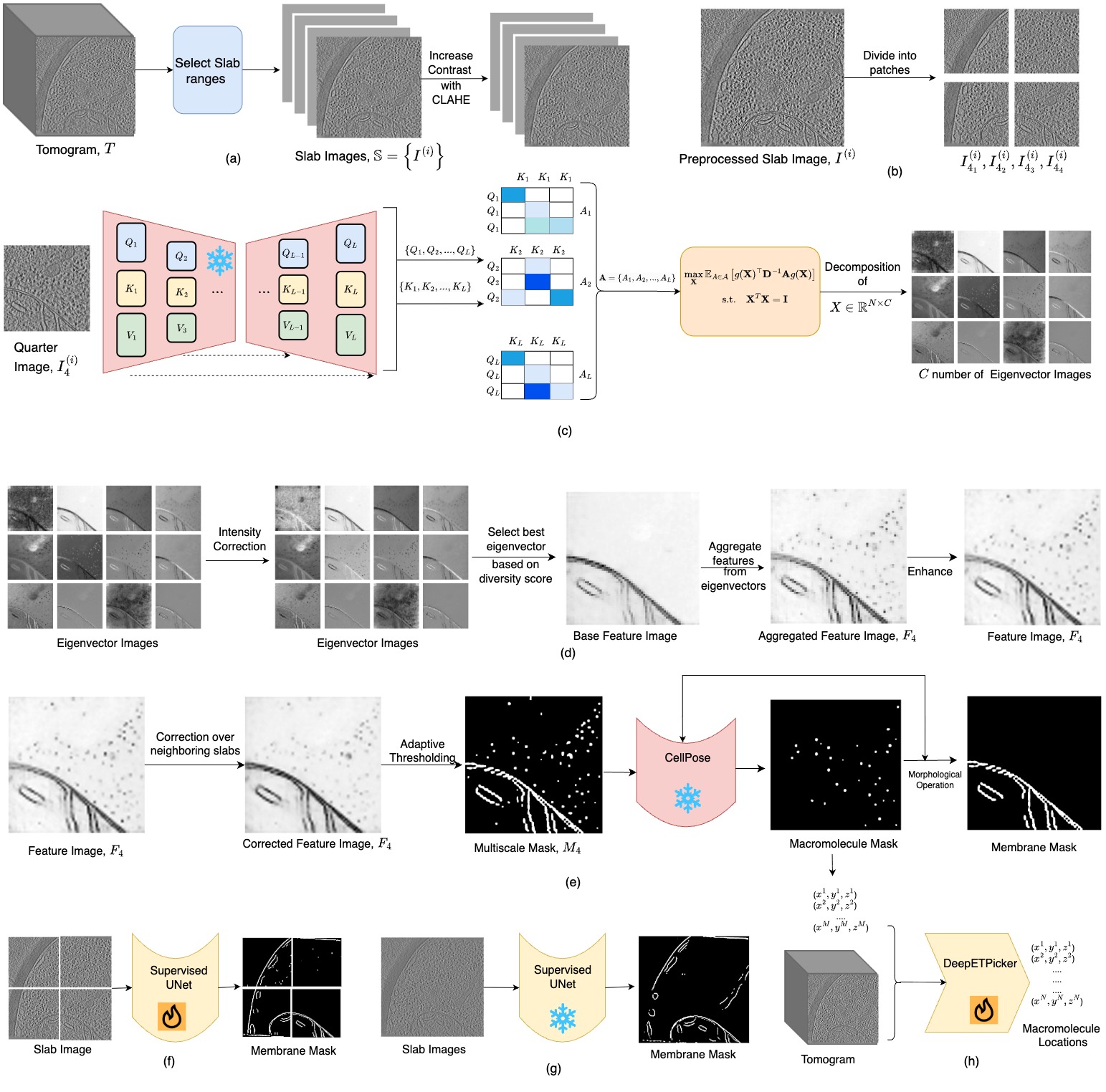
Unsupervised Multi-scale Segmentation of Cellular cryo-electron Tomograms with Stable Diffusion Foundation Model
We introduce an unsupervised approach for segmenting multiscale subcellular objects in 3D volumetric cryo-electron tomography (cryo-ET) images, addressing key challenges such as large data volumes, low signal-to-noise ratios, and the heterogeneity of subcellular shapes and sizes. The method requires users to select a small number of slabs from a few representative tomograms in the dataset. It leverages features extracted from all layers of a Stable Diffusion foundation model, followed by a novel heuristic- based feature aggregation strategy. Segmentation masks are generated using adaptive thresholding, refined with CellPose to split composite regions, and then utilized as pseudo-ground truth for training deep learning models. This fully automated, data-driven framework enables the mining of multi-scale subcellular patterns, paving the way for accelerated biological discoveries from large-scale cellular cryo-ET datasets.
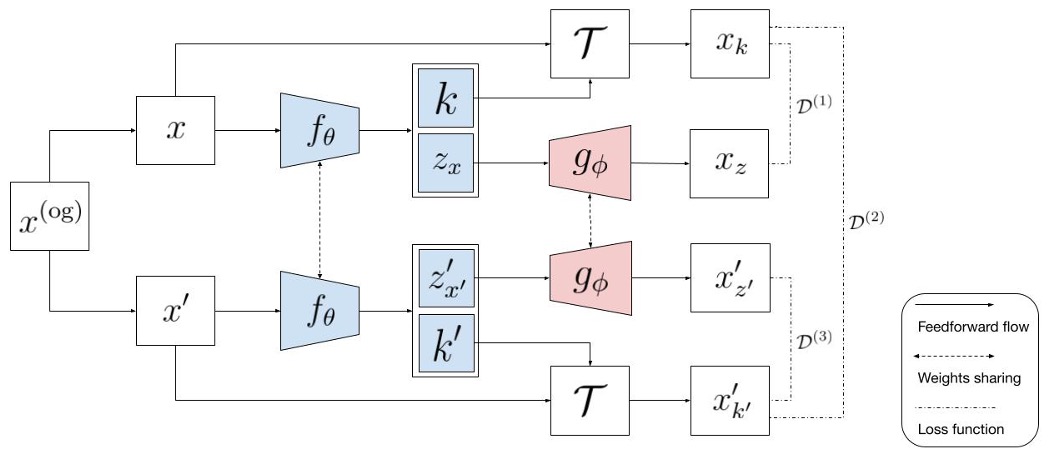
A Generic Unsupervised Approach for Disentangling Semantic Content from Parameterized Transformations.
In many real-life image analysis applications, particularly in biomedical research domains, the objects of interest undergo multiple transformations that alter their visual properties while keeping the semantic content unchanged. Disentangling images into semantic content factors and transformations can provide significant benefits for many domain-specific image analysis tasks. To this end, we propose a generic unsupervised framework, Harmony, that simultaneously and explicitly disentangles semantic content from multiple parameterized transformations. Harmony leverages a simple cross-contrastive learning framework with multiple explicitly parameterized latent representations to disentangle content from transformations. To demonstrate the efficacy of Harmony, we apply it to disentangle image semantic content from several parameterized transformations (rotation, translation, scaling, and contrast). Harmony achieves significantly improved disentanglement over the baseline models on several image datasets of diverse domains. With such disentanglement, Harmony is demonstrated to incentivize bioimage analysis research by modeling structural heterogeneity of macromolecules from cryo-ET images and learning transformation-invariant representations of protein particles from single-particle cryo-EM images. Therefore, Harmony is generalizable to many other imaging domains and can potentially be extended to domains beyond imaging as well.
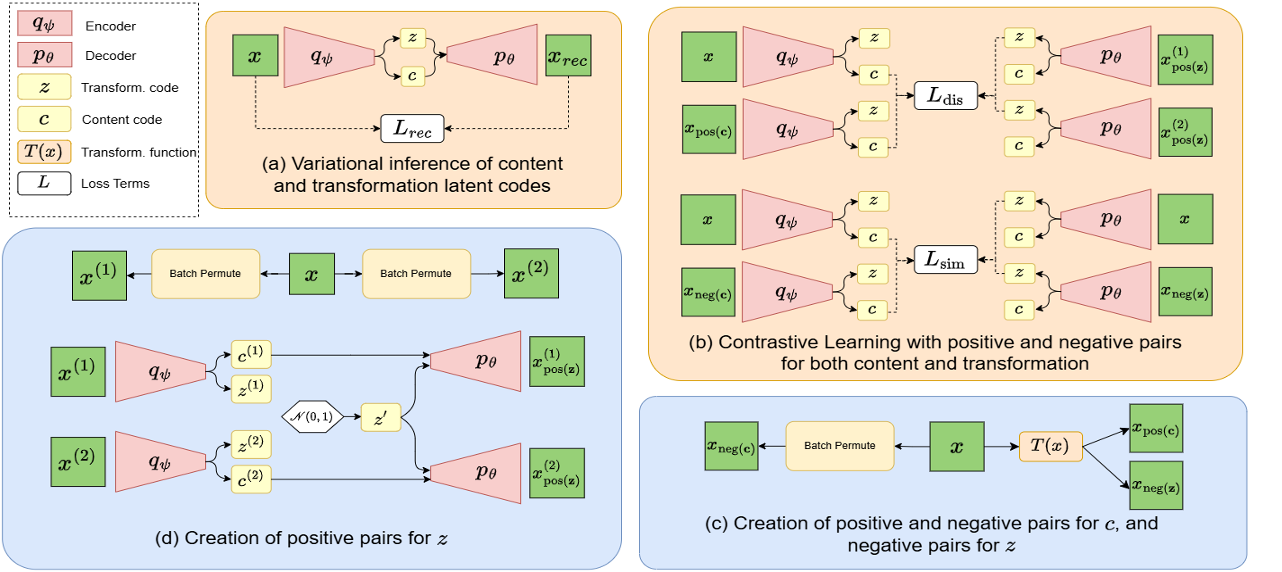
Unsupervised Identification of Protein Compositions and Conformations via Implicit Content-Transformation Disentanglement
Identifying different protein compositions and conformations from microscopic images of protein mixtures is a challenging open problem. We address this through disentangled representation learning, where separating protein compositions and conformations in an intermediate latent space enables accurate identification. By modeling compositions and conformations as content and transformation, the task can be reduced to disentangling content and transformation. The existing disentangling methods require an explicit parametric form for the transformation, which is unavailable for conformation, making these methods unsuitable. To overcome this limitation, we propose DualContrast, a novel contrastive learning-based method that implicitly parameterizes and disentangles both transformation and content. DualContrast achieves this by generating positive and negative pairs for content and transformation in both data and latent spaces. We demonstrate that existing self-supervised approaches fail under similar implicit parameterization, underscoring the necessity of our method. Through extensive experiments on 3D microscopic images of protein mixtures and additional shape-focused datasets beyond microscopy, we validate our claims and demonstrate the first in-principle fully unsupervised identification of different protein compositions and conformations in 3D microscopic images.
Teaching Experiences
Carnegie Mellon University- GTA
- CMU02-620 Machine Learning for Scientists : A graduate level machine Learning course designed for Masters students in automated science and computational biology in SCS.
- CMU02-740 Bioimage Informatics : A graduate level course on biological image processing designed for Masters students in automated science and computational biology in SCS.
East West University- Lecturer
- CSE498 Social and Professional Issues in Computing
- CSE103 Structured Programming
- CSE101 Computer Fundamentals
- CSE350 Data Communications
- CSE106 Discrete Mathematics
Awards & Honors
- CPCB Outstanding Research Accomplishment Award
- CMLH Fellowshiop in Digital Health 2023 news
- Deans List Award and University Merit Scholarship
- 1st Place - Poster Presentation, International Conference on Networking, Systems and Security (4th NSysS 2017) pdf
- 1st Place - Hackathon for environmental migrants in Bangladesh arranged by Wageningen University, Netherlands. report
- 2nd Place - Bracathon 2017 by BRAC
- 3rd Place - National Hackathon 2016 by ICT Division.
-
3rd Place - BUET Website Design Competition by IICT, BUET
Activities & Workshop
-
Served as a reviewer in IEEE Computer Vision and Pattern Recognition (CVPR) 2022, 2023, 2025, International Conference on Computer Vision (ICCV) 2023, 2025, European Conference on Computer Vision (ECCV) 2022, 2024, and AAAI Conference 2023, 2024.
-
Works as a mentor in CMU AI Mentoring Program, where I mentor CMU undergraduate students coming from underrepresented communities interested in AI research.
-
Worked as a moderator of East West University Electronics, Programming and Robotics Club. (Jan 2020- Dec 2020)
-
Designed and developed a responsive website for International Conference on Networking, Systems and Security(NSysS) jointly with Ajoy Das, under supervision of Dr. Rifat Shahriyar. Website Link.
-
Participated in a workshop on ``Reverse Engineering" arranged by ICT Division, Bangladesh Government. A team consisting of 18 members from CSE, BUET was provided with the opportunity to attend this workshop. The workshop was conducted by Dr. Desmond Devendran.
-
Participated in reviewing National ICT books as a team member of CSE, BUET.
-
Actively worked as an organizer of BUET CSE FEST 2018.
Mini-Projects
Design of Phase-separated Protein Sequences using Adaptive Sampling and Active Learning
We address the problem of in silico protein design with a high propensity for liquid-liquid phase separation (LLPS) and droplet formation. Recently, there has been a surge in computational protein design methods that exhibit certain functions or structures. Moreover, no current method explicitly addresses the problem of computationally designing proteins with a high propensity for phase separation. To this end, we, for the first time, developed an adaptive sampling-based approach for in silico phase-separation protein design. Our method consists of multiple components, including a relaxed “energy" based sequence generator, a biochemical condition-aware attention-neural network-based surrogate model, a Bayesian acquisition function, and its optimizer. We demonstrate that our pipeline effectively generates in silico proteins with a high propensity for droplet formation in LLPS experiments, which outperforms other design methods.
Edge prediction: Predicting Edge in Academic Citation Networks
This project attempts to assess the performance of various methods for predicting the citation of academic articles. Many researchers have sought to predict the future citation of new articles, and this interest has resulted in researchers using various machine learning methods for prediction. Our work asks a slightly different but related question. Given an article, how likely is it to cite another particular article? For our specific task, we found that sophisticated graph structure-based model does not achieve very promising performance. To this end, we developed an intelligent and novel feature engineering pipeline that could generate highly accurate predictions with relatively simpler models. We achieved around 95% F1 score with random forest classifier with our engineered features, which largely outperformed the graph neural network-based model.
Pytorch Autograd Implementation of OpenMM local energy minimizer
In this project, we implemented the openmm local energy minimizer (that is used to minimize the free energy of protein in protein dynamics) using pytorch. We extended the autograd mechanics of pytorch for a custom backpropagation where in the forward pass the energy is calculated and in the backward pass, each atom's coordinate is updated according to the energy gradients. This work was done under supervision of Prof. David Koes.
Predicting age from lung single cell data
In this project, we have analyzed the scRNA-seq data for 28 control patients to predict biological age from them. We tested with different machine learning approaches along with popular feature extraction methods and reported the results.This project was done as a lab rotation work with Prof. Ziv Bar-Joseph.
Bangla to English Machine Translation Using Seq2seq Model with Attention Mechanism
In this term project, we did an experiment on Neural Machine Translation(NMT) for Bangla to English Translation. We used a moderate size dataset containing 4379 sentence translations from English to Bangla. We used seq2seq encoder-decoder model containing Word2Vec and LSTMs with and without attention for small epochs. With finely tuned hyperparameters, we observed that using Bahdanau's attention with the vanilla encoder-decoder model improves the BLEU score for Bangla to English translation.
Contact
7513 Gates and Hillman Centers
Carnegie Mellon University
5000 Forbes Avenue
Pittsburgh, PA 15213, USA